from pathlib import Path
from functools import reduceCluster Gender Changes
This notebook audits for significant changes in cluster gender annotations, to allow us to detect the significance of shifts over time. It depends on the aligned cluster identities in isbn-version-clusters.parquet.
import pandas as pd
import polars as pl
import numpy as np
import matplotlib.pyplot as plt
import seaborn as snsLoad Data
Define the versions we care about:
versions = ['pgsql', '2022-03-2.0', '2022-07', '2022-10', '2022-11-2.1', '2023-07', '2025-06-3.0', 'current']Load the aligned ISBNs:
isbn_clusters = pd.read_parquet('isbn-version-clusters.parquet')
isbn_clusters.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 44866744 entries, 0 to 44866743
Data columns (total 10 columns):
# Column Dtype
--- ------ -----
0 isbn object
1 isbn_id int32
2 current float64
3 2025-06-3.0 float64
4 2023-07 float64
5 2022-11-2.1 float64
6 2022-10 float64
7 2022-07 float64
8 2022-03-2.0 float64
9 pgsql float64
dtypes: float64(8), int32(1), object(1)
memory usage: 3.2+ GBDifferent Genders
How many clusters changed gender?
To get started, we need a list of genders in order.
genders = [
'ambiguous', 'female', 'male', 'unknown',
'no-author-rec', 'no-book-author', 'no-book', 'absent'
]Let’s make a function to read gender info:
def read_gender(path, map_file=None):
cg = pl.scan_parquet(path)
cg = cg.select([
pl.col('cluster').cast(pl.Int32),
pl.when(pl.col('gender') == pl.lit('no-loc-author'))
.then(pl.lit('no-book-author'))
.when(pl.col('gender') == pl.lit('no-viaf-author'))
.then(pl.lit('no-author-rec'))
.otherwise(pl.col('gender'))
.cast(pl.Categorical)
.alias('gender')
])
if map_file is not None:
map = pl.scan_parquet(map_file)
cg = cg.join(map, on='cluster', how='left')
cg = cg.select([
pl.col('common').alias('cluster'),
pl.col('gender')
])
return cgRead each data source’s gender info and map to common cluster IDs:
gender_cc = {
v: read_gender(f'{v}/cluster-genders.parquet', f'{v}/cluster-map.parquet')
for v in versions if v != 'current'
}
gender_cc['current'] = read_gender('../book-links/cluster-genders.parquet')Set up a sequence of frames for merging:
to_merge = [
gender_cc[v].select([
pl.col('cluster'),
pl.col('gender').alias(v)
]).unique()
for v in versions
]Merge and collect results:
cluster_genders = reduce(lambda df1, df2: df1.join(df2, on='cluster', how='outer_coalesce'), to_merge)
cluster_genders = cluster_genders.collect()/tmp/ipykernel_4174369/2981784951.py:1: DeprecationWarning: Use of `how='outer_coalesce'` should be replaced with `how='full', coalesce=True`.
cluster_genders = reduce(lambda df1, df2: df1.join(df2, on='cluster', how='outer_coalesce'), to_merge)For unclear reasons, a few versions have a null cluster. Drop that.
cluster_genders = cluster_genders.filter(cluster_genders['cluster'].is_not_null())Now we will convert to Pandas and fix missing values:
cluster_genders = cluster_genders.to_pandas().set_index('cluster')Now we’ll unify the categories and their orders:
cluster_genders = cluster_genders.apply(lambda vdf: vdf.cat.set_categories(genders, ordered=True))
cluster_genders.fillna('absent', inplace=True)
cluster_genders.head()| pgsql | 2022-03-2.0 | 2022-07 | 2022-10 | 2022-11-2.1 | 2023-07 | 2025-06-3.0 | current | |
|---|---|---|---|---|---|---|---|---|
| cluster | ||||||||
| 101559774 | absent | absent | absent | absent | absent | absent | absent | no-book-author |
| 452208798 | no-book-author | no-book | no-book | no-book-author | no-book-author | no-book-author | no-book-author | no-book-author |
| 127012979 | absent | absent | no-book-author | no-book-author | no-book-author | no-book-author | no-book-author | no-book-author |
| 117407548 | no-book-author | no-book-author | no-book-author | no-book-author | no-book-author | no-book-author | no-book-author | no-book-author |
| 102777110 | female | female | female | female | female | female | female | female |
Let’s save this file for further analysis:
cluster_genders.to_parquet('cluster-version-genders.parquet', compression='zstd')PostgreSQL to Current
Now we are ready to actually compare cluster genders across categories. Let’s start by comparing original data (PostgreSQL) to current:
ct = cluster_genders[['pgsql', 'current']].value_counts().unstack()
ct = ct.reindex(labels=genders, columns=genders)
ct| current | ambiguous | female | male | unknown | no-author-rec | no-book-author | no-book | absent |
|---|---|---|---|---|---|---|---|---|
| pgsql | ||||||||
| ambiguous | 95825.0 | 4815.0 | 11222.0 | 2398.0 | 1660.0 | 3175.0 | 4.0 | 5109.0 |
| female | 17856.0 | 1106202.0 | 1224.0 | 19534.0 | 9317.0 | 256.0 | 31.0 | 39691.0 |
| male | 30709.0 | 3094.0 | 3472004.0 | 28199.0 | 30960.0 | 808.0 | 155.0 | 79287.0 |
| unknown | 3222.0 | 130124.0 | 230286.0 | 1501820.0 | 19732.0 | 21.0 | 12.0 | 15158.0 |
| no-author-rec | 10753.0 | 60165.0 | 332493.0 | 230073.0 | 1385651.0 | 1097.0 | 125.0 | 15337.0 |
| no-book-author | 9084.0 | 114768.0 | 222317.0 | 132370.0 | 213811.0 | 2445284.0 | 898251.0 | 278188.0 |
| no-book | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| absent | 130365.0 | 1087795.0 | 2667206.0 | 1177309.0 | 4962030.0 | 17648931.0 | 728440.0 | 358958.0 |
ctf = ct.divide(ct.sum(axis='columns'), axis='rows')
def style_row(row):
styles = []
for col, val in zip(row.index, row.values):
if col == row.name:
styles.append('font-weight: bold')
elif val > 0.1:
styles.append('color: red')
else:
styles.append(None)
return styles
ctf.style.apply(style_row, 'columns')| current | ambiguous | female | male | unknown | no-author-rec | no-book-author | no-book | absent |
|---|---|---|---|---|---|---|---|---|
| pgsql | ||||||||
| ambiguous | 0.771488 | 0.038766 | 0.090348 | 0.019306 | 0.013365 | 0.025562 | 0.000032 | 0.041133 |
| female | 0.014953 | 0.926381 | 0.001025 | 0.016359 | 0.007802 | 0.000214 | 0.000026 | 0.033239 |
| male | 0.008424 | 0.000849 | 0.952482 | 0.007736 | 0.008493 | 0.000222 | 0.000043 | 0.021751 |
| unknown | 0.001695 | 0.068473 | 0.121179 | 0.790276 | 0.010383 | 0.000011 | 0.000006 | 0.007976 |
| no-author-rec | 0.005282 | 0.029555 | 0.163332 | 0.113019 | 0.680677 | 0.000539 | 0.000061 | 0.007534 |
| no-book-author | 0.002106 | 0.026603 | 0.051533 | 0.030683 | 0.049561 | 0.566816 | 0.208214 | 0.064484 |
| no-book | nan | nan | nan | nan | nan | nan | nan | nan |
| absent | 0.004533 | 0.037822 | 0.092737 | 0.040934 | 0.172526 | 0.613640 | 0.025327 | 0.012481 |
Most of the change is coming from clusters absent in the original but present in the new.
There are also quite a few that had no book author in PGSQL, but no book in the current data - not sure what’s up with that. Let’s look at more crosstabs.
def gender_crosstab(old, new, fractional=True):
ct = cluster_genders[[old, new]].value_counts().unstack()
ct = ct.reindex(labels=genders, columns=genders)
if fractional:
ctf = ct.divide(ct.sum(axis='columns'), axis='rows')
return ctf
else:
return ctdef plot_gender(set):
cluster_genders[set].value_counts().sort_index().plot.barh()
plt.title(f'Gender Distribution in {set}')PostgreSQL to March 2022 (2.0 release)
This marks the change from PostgreSQL to pure-Rust.
ct = gender_crosstab('pgsql', '2022-03-2.0')
ct.style.apply(style_row, 'columns')| 2022-03-2.0 | ambiguous | female | male | unknown | no-author-rec | no-book-author | no-book | absent |
|---|---|---|---|---|---|---|---|---|
| pgsql | ||||||||
| ambiguous | 0.977924 | 0.002955 | 0.013928 | 0.000636 | 0.000878 | nan | nan | 0.003679 |
| female | 0.002190 | 0.993945 | 0.000001 | 0.000301 | 0.000430 | 0.000003 | 0.000005 | 0.003125 |
| male | 0.000591 | 0.000000 | 0.995938 | 0.000528 | 0.000796 | 0.000002 | 0.000014 | 0.002131 |
| unknown | 0.000043 | 0.002759 | 0.005304 | 0.988899 | 0.001953 | 0.000001 | 0.000003 | 0.001039 |
| no-author-rec | 0.000104 | 0.007919 | 0.049969 | 0.031480 | 0.908599 | nan | 0.000008 | 0.001921 |
| no-book-author | 0.000002 | 0.000051 | 0.000200 | 0.000108 | 0.000051 | 0.649173 | 0.335017 | 0.015397 |
| no-book | nan | nan | nan | nan | nan | nan | nan | nan |
| absent | 0.000006 | 0.000046 | 0.000239 | 0.000112 | 0.000167 | 0.001884 | 0.000066 | 0.997481 |
This is where we change from no-book-author to no-book for a bunch of books; otherwise things are pretty consistent. This major change is likely a result of changes that count more books and book clusters - we had some inner joins in the PostgreSQL version that were questionable, and in particular we didn’t really cluster solo ISBNs but now we do. But now, if we have a solo ISBN from rating data, it gets a cluster with no book record instead of being excluded from the clustering.
Let’s look at the distribution of statuses for each, starting with PostgreSQL:
plot_gender('pgsql')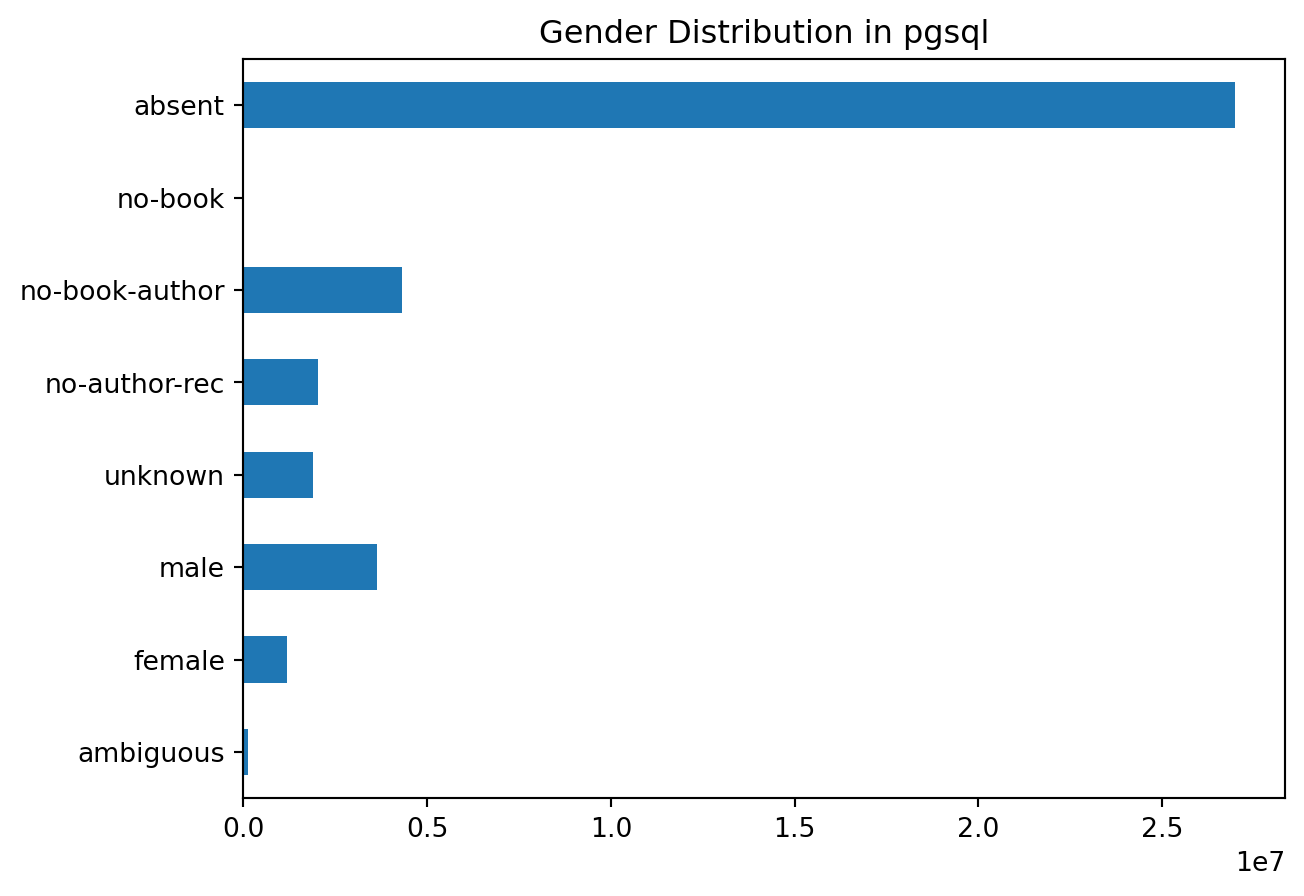
And the Rust version:
plot_gender('2022-03-2.0')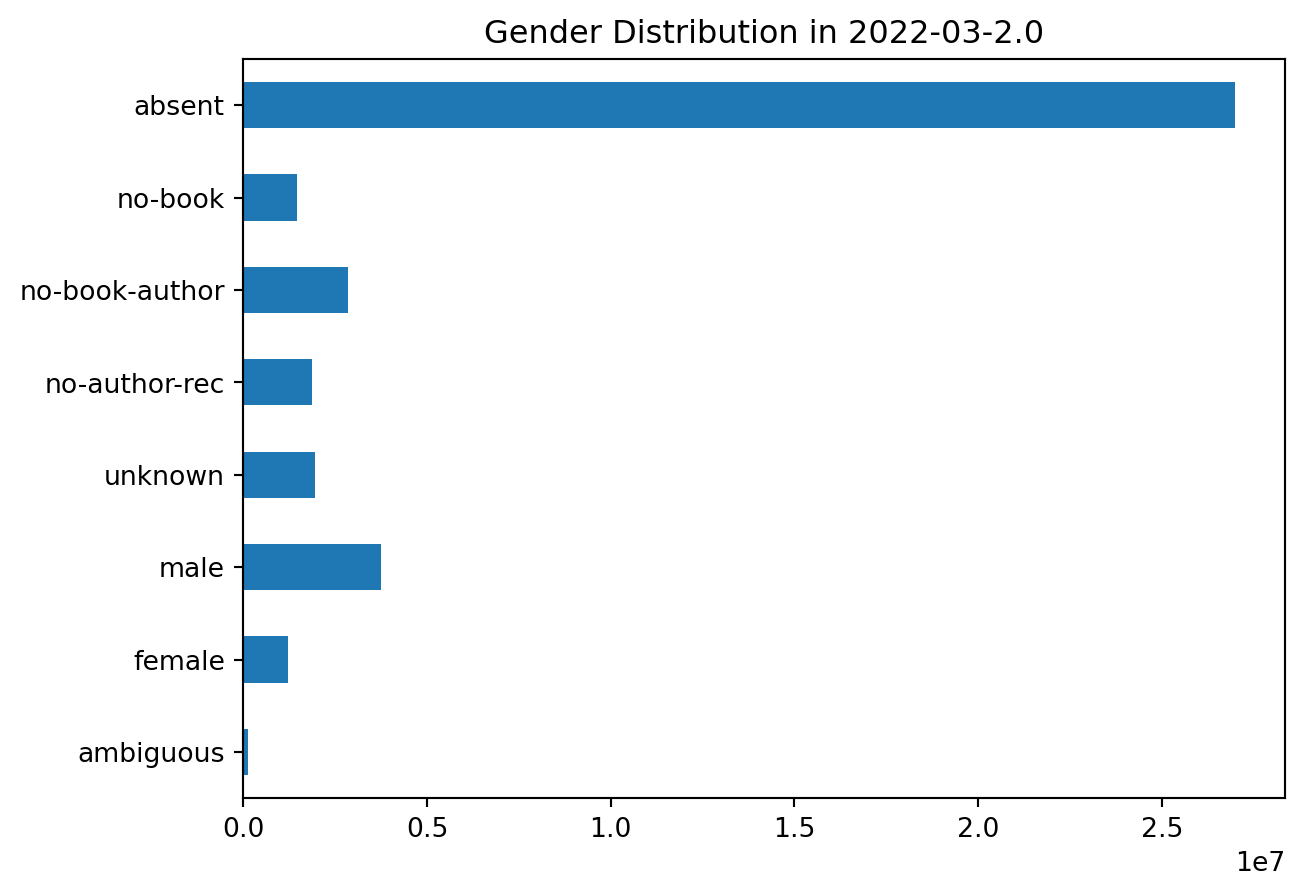
March to July 2022
We updated a lot of data files and changed the name and ISBN parsing logic.
ct = gender_crosstab('2022-03-2.0', '2022-07')
ct.style.apply(style_row, 'columns')| 2022-07 | ambiguous | female | male | unknown | no-author-rec | no-book-author | no-book | absent |
|---|---|---|---|---|---|---|---|---|
| 2022-03-2.0 | ||||||||
| ambiguous | 0.836228 | 0.035602 | 0.083587 | 0.014814 | 0.004862 | 0.000087 | 0.000016 | 0.024805 |
| female | 0.010065 | 0.963186 | 0.000487 | 0.007911 | 0.001253 | 0.000002 | 0.000012 | 0.017085 |
| male | 0.006706 | 0.000646 | 0.974636 | 0.003702 | 0.001364 | 0.000079 | 0.000014 | 0.012852 |
| unknown | 0.001901 | 0.040305 | 0.092947 | 0.856051 | 0.003411 | 0.000200 | nan | 0.005186 |
| no-author-rec | 0.003536 | 0.020630 | 0.108694 | 0.101641 | 0.762110 | 0.000009 | 0.000037 | 0.003344 |
| no-book-author | 0.002056 | 0.030443 | 0.057337 | 0.035301 | 0.056239 | 0.809983 | 0.000007 | 0.008635 |
| no-book | 0.000159 | 0.002344 | 0.005280 | 0.002742 | 0.004765 | 0.000634 | 0.980665 | 0.003411 |
| absent | 0.002109 | 0.018914 | 0.040477 | 0.014926 | 0.059121 | 0.003270 | 0.000001 | 0.861182 |
Mostly fine; some more are resolved, existing resolutions are pretty consistent.
plot_gender('2022-07')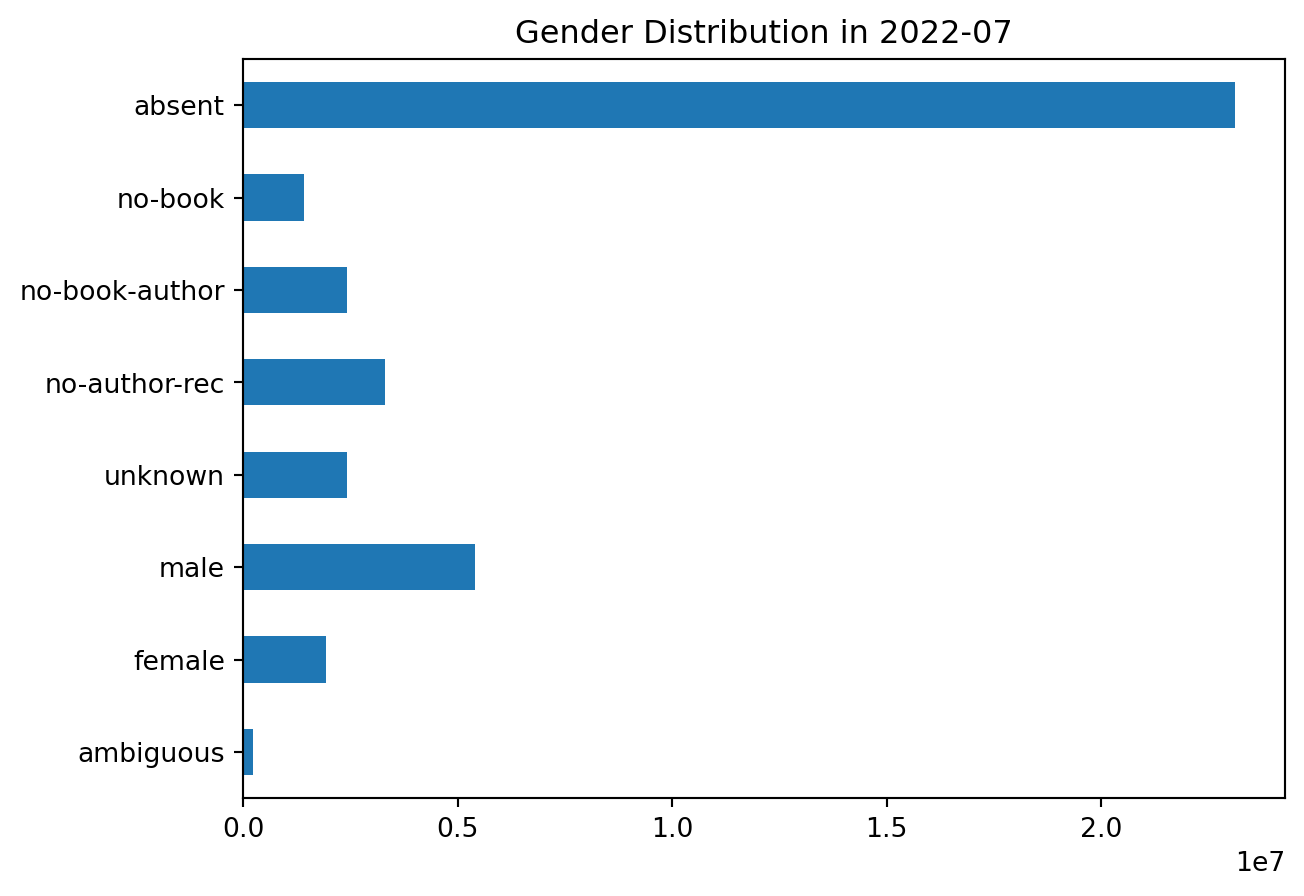
July 2022 to Oct. 2022
We changed from DataFusion to Polars and made further ISBN and name parsing changes.
ct = gender_crosstab('2022-07', '2022-10')
ct.style.apply(style_row, 'columns')| 2022-10 | ambiguous | female | male | unknown | no-author-rec | no-book-author | no-book | absent |
|---|---|---|---|---|---|---|---|---|
| 2022-07 | ||||||||
| ambiguous | 0.989385 | 0.004969 | 0.003626 | 0.000336 | 0.001647 | nan | nan | 0.000036 |
| female | nan | 0.995099 | nan | 0.000361 | 0.004471 | nan | nan | 0.000070 |
| male | 0.000001 | nan | 0.994580 | 0.000431 | 0.004975 | 0.000000 | nan | 0.000012 |
| unknown | nan | nan | nan | 0.995467 | 0.004492 | nan | nan | 0.000042 |
| no-author-rec | nan | 0.000001 | 0.000003 | 0.000005 | 0.999823 | 0.000131 | nan | 0.000038 |
| no-book-author | nan | 0.000000 | 0.000001 | 0.000000 | 0.000000 | 0.996612 | nan | 0.003386 |
| no-book | 0.000004 | 0.000089 | 0.000035 | 0.000069 | 0.000094 | 0.198066 | 0.670216 | 0.131427 |
| absent | nan | nan | nan | nan | nan | nan | nan | 1.000000 |
plot_gender('2022-10')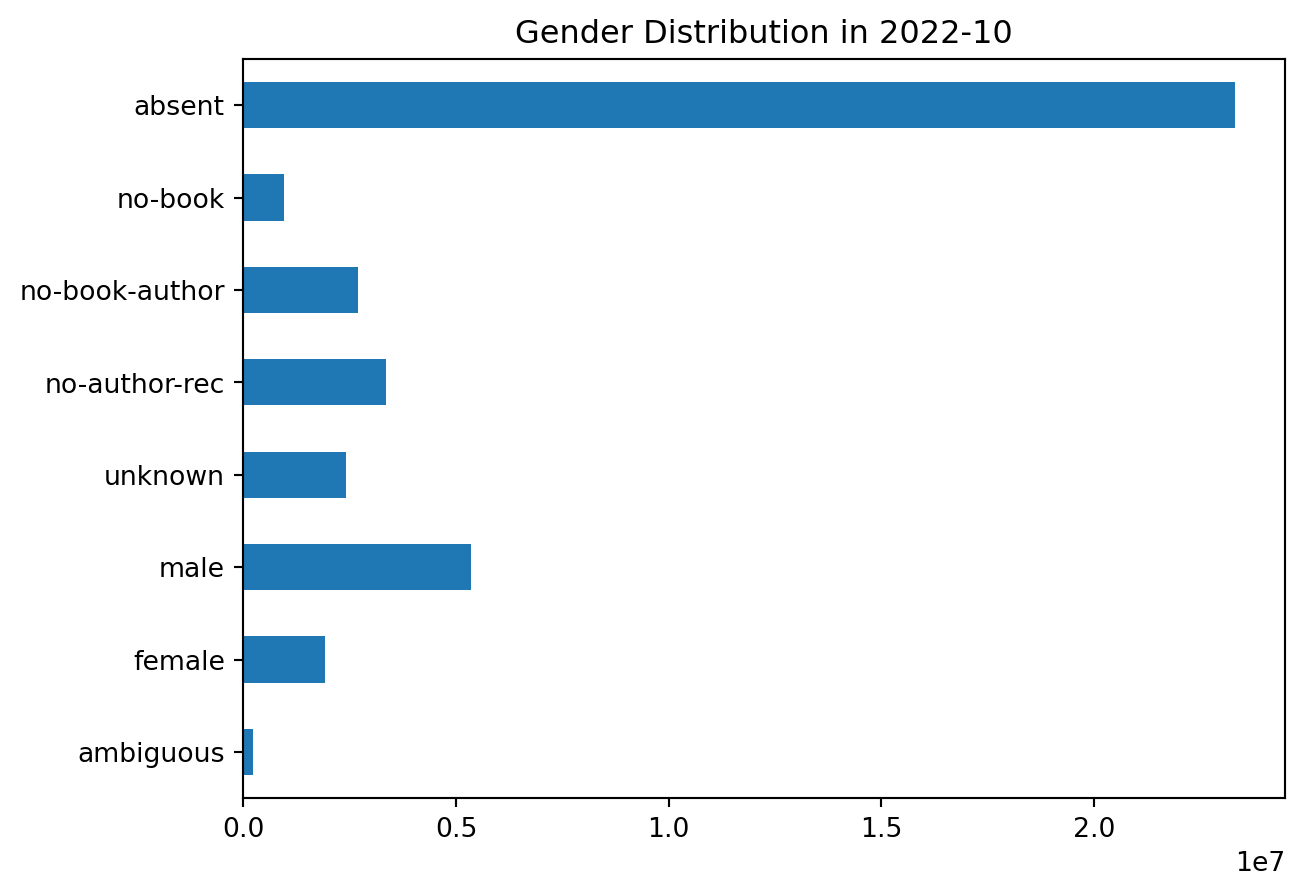
Oct. 2022 to release 2.1 (Nov. 2022)
We added support for GoodReads CSV data and the Amazon 2018 rating CSV files.
ct = gender_crosstab('2022-10', '2022-11-2.1')
ct.style.apply(style_row, 'columns')| 2022-11-2.1 | ambiguous | female | male | unknown | no-author-rec | no-book-author | no-book | absent |
|---|---|---|---|---|---|---|---|---|
| 2022-10 | ||||||||
| ambiguous | 0.999995 | nan | nan | nan | nan | nan | nan | 0.000005 |
| female | nan | 0.999999 | nan | nan | nan | nan | nan | 0.000001 |
| male | nan | nan | 0.999999 | nan | nan | nan | nan | 0.000001 |
| unknown | nan | nan | nan | 0.999998 | nan | nan | nan | 0.000002 |
| no-author-rec | nan | nan | nan | nan | 0.999999 | nan | nan | 0.000001 |
| no-book-author | nan | nan | nan | 0.000000 | nan | 0.999982 | nan | 0.000017 |
| no-book | nan | nan | nan | nan | nan | nan | 1.000000 | nan |
| absent | nan | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000003 | 0.031489 | 0.968508 |
plot_gender('2022-11-2.1')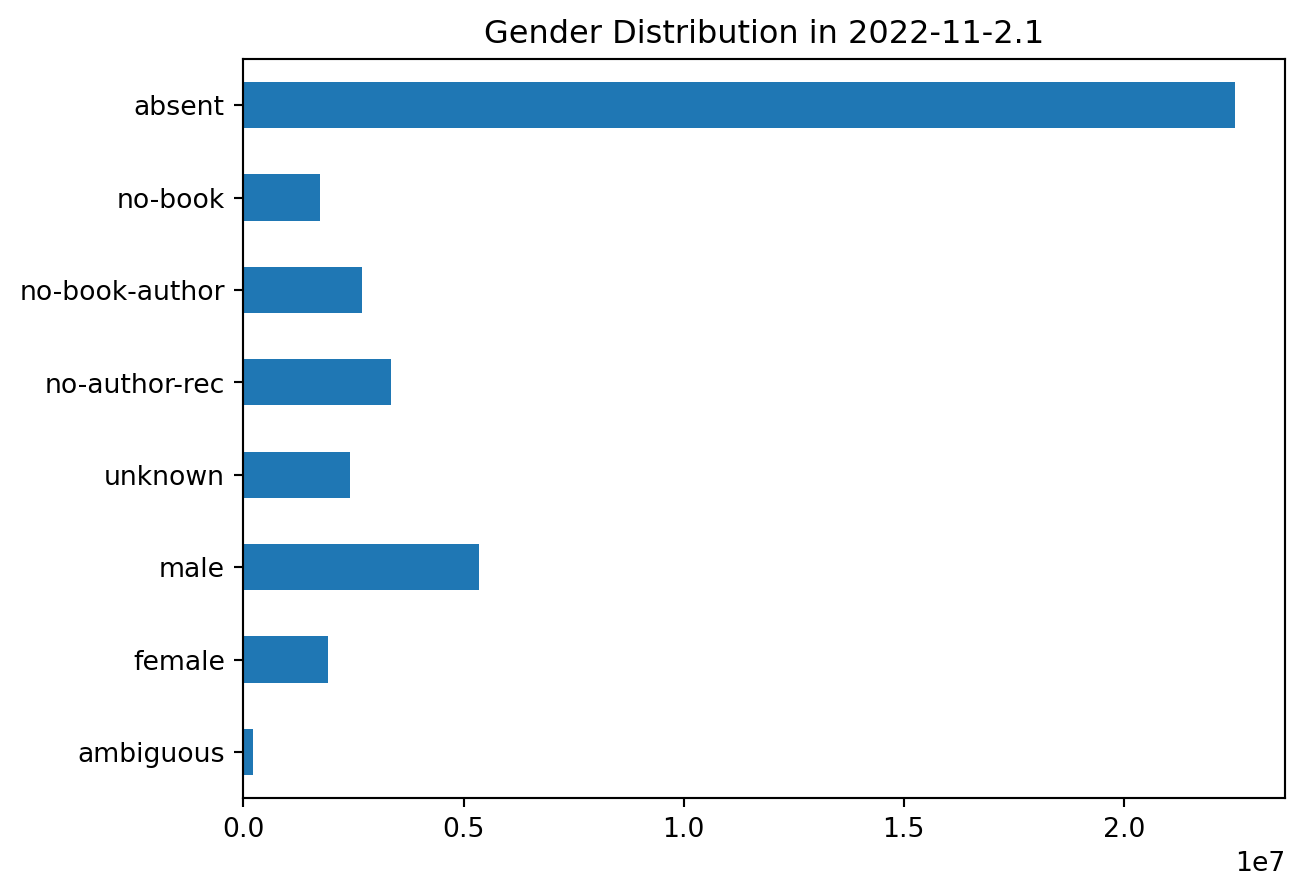
Release 2.1 to Jul. 2023
We updated OpenLibrary and VIAF, and made some technical changes.
ct = gender_crosstab('2022-11-2.1', '2023-07')
ct.style.apply(style_row, 'columns')| 2023-07 | ambiguous | female | male | unknown | no-author-rec | no-book-author | no-book | absent |
|---|---|---|---|---|---|---|---|---|
| 2022-11-2.1 | ||||||||
| ambiguous | 0.879459 | 0.006921 | 0.018915 | 0.001573 | 0.000239 | 0.074744 | 0.000009 | 0.018140 |
| female | 0.004892 | 0.975566 | 0.000307 | 0.004275 | 0.000396 | 0.000482 | 0.000002 | 0.014081 |
| male | 0.002249 | 0.000044 | 0.986211 | 0.001428 | 0.000497 | 0.001463 | 0.000005 | 0.008105 |
| unknown | 0.000266 | 0.019105 | 0.032232 | 0.945164 | 0.001086 | 0.000425 | 0.000000 | 0.001721 |
| no-author-rec | 0.000277 | 0.002896 | 0.007170 | 0.009781 | 0.975855 | 0.000360 | 0.000004 | 0.003658 |
| no-book-author | 0.000408 | 0.006223 | 0.011060 | 0.004081 | 0.006726 | 0.967637 | 0.000001 | 0.003864 |
| no-book | 0.000363 | 0.005640 | 0.011780 | 0.007411 | 0.024719 | 0.003812 | 0.940827 | 0.005448 |
| absent | 0.002871 | 0.019474 | 0.052188 | 0.024984 | 0.118197 | 0.013907 | 0.000001 | 0.768377 |
plot_gender('2023-07')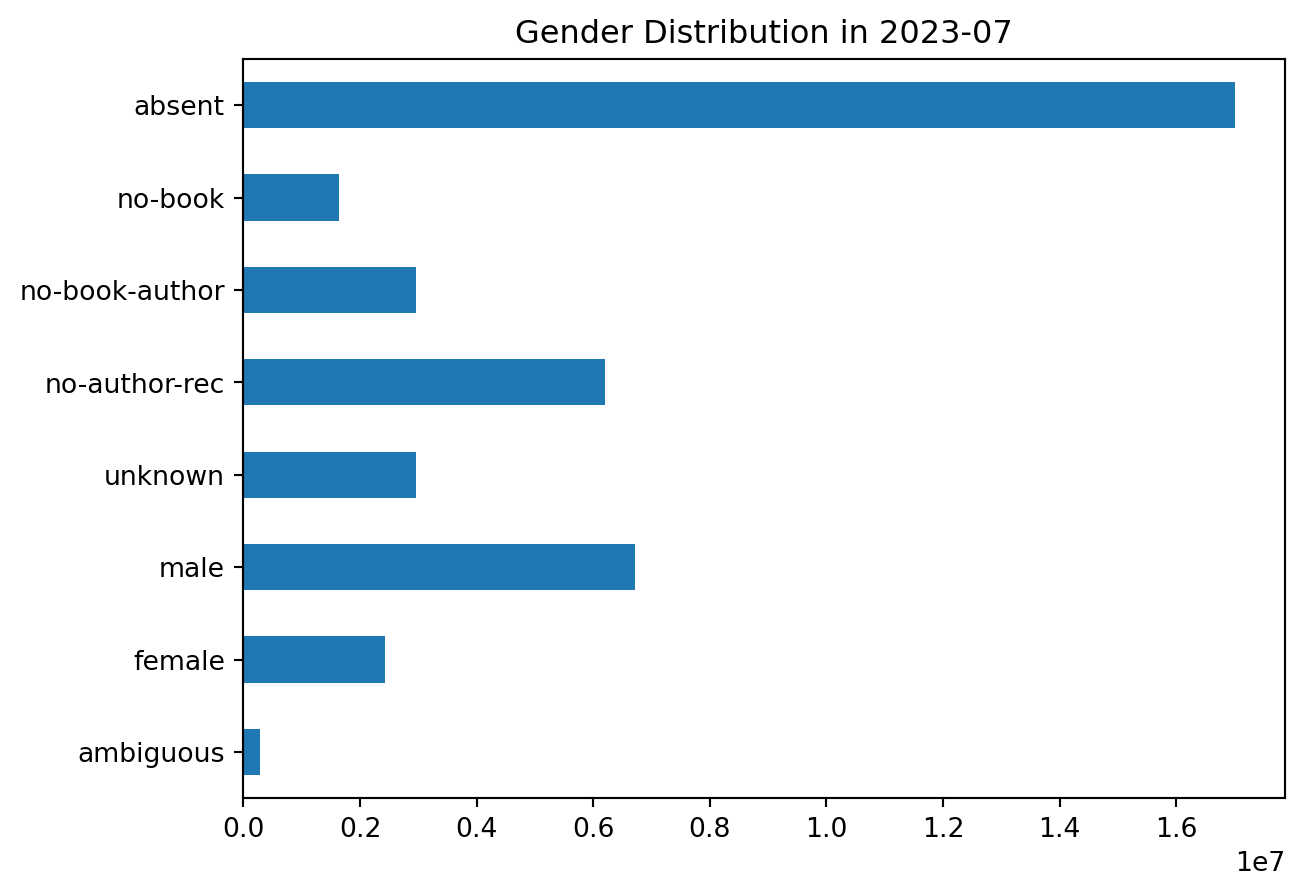
Jul. 2023 to 3.0 (June 2025)
Mostly technical changes, along with updated OpenLibrary an dVIAF
ct = gender_crosstab('2023-07', '2025-06-3.0')
ct.style.apply(style_row, 'columns')| 2025-06-3.0 | ambiguous | female | male | unknown | no-author-rec | no-book-author | no-book | absent |
|---|---|---|---|---|---|---|---|---|
| 2023-07 | ||||||||
| ambiguous | 0.916513 | 0.015526 | 0.040841 | 0.004112 | 0.000437 | 0.005387 | nan | 0.017185 |
| female | 0.003798 | 0.962122 | 0.000154 | 0.023668 | 0.000251 | 0.000479 | 0.000000 | 0.009528 |
| male | 0.002358 | 0.000091 | 0.980826 | 0.010716 | 0.000521 | 0.000440 | 0.000001 | 0.005047 |
| unknown | 0.000162 | 0.021824 | 0.023425 | 0.949053 | 0.002647 | 0.000014 | nan | 0.002876 |
| no-author-rec | 0.000114 | 0.001472 | 0.002637 | 0.005071 | 0.952103 | 0.000162 | 0.000015 | 0.038426 |
| no-book-author | 0.000084 | 0.000694 | 0.001612 | 0.000504 | 0.000704 | 0.994608 | 0.000000 | 0.001794 |
| no-book | 0.000037 | 0.000464 | 0.000953 | 0.000865 | 0.003197 | 0.001035 | 0.992922 | 0.000525 |
| absent | 0.000476 | 0.004847 | 0.013392 | 0.006382 | 0.036988 | 0.003447 | 0.000001 | 0.934466 |
plot_gender('2025-06-3.0')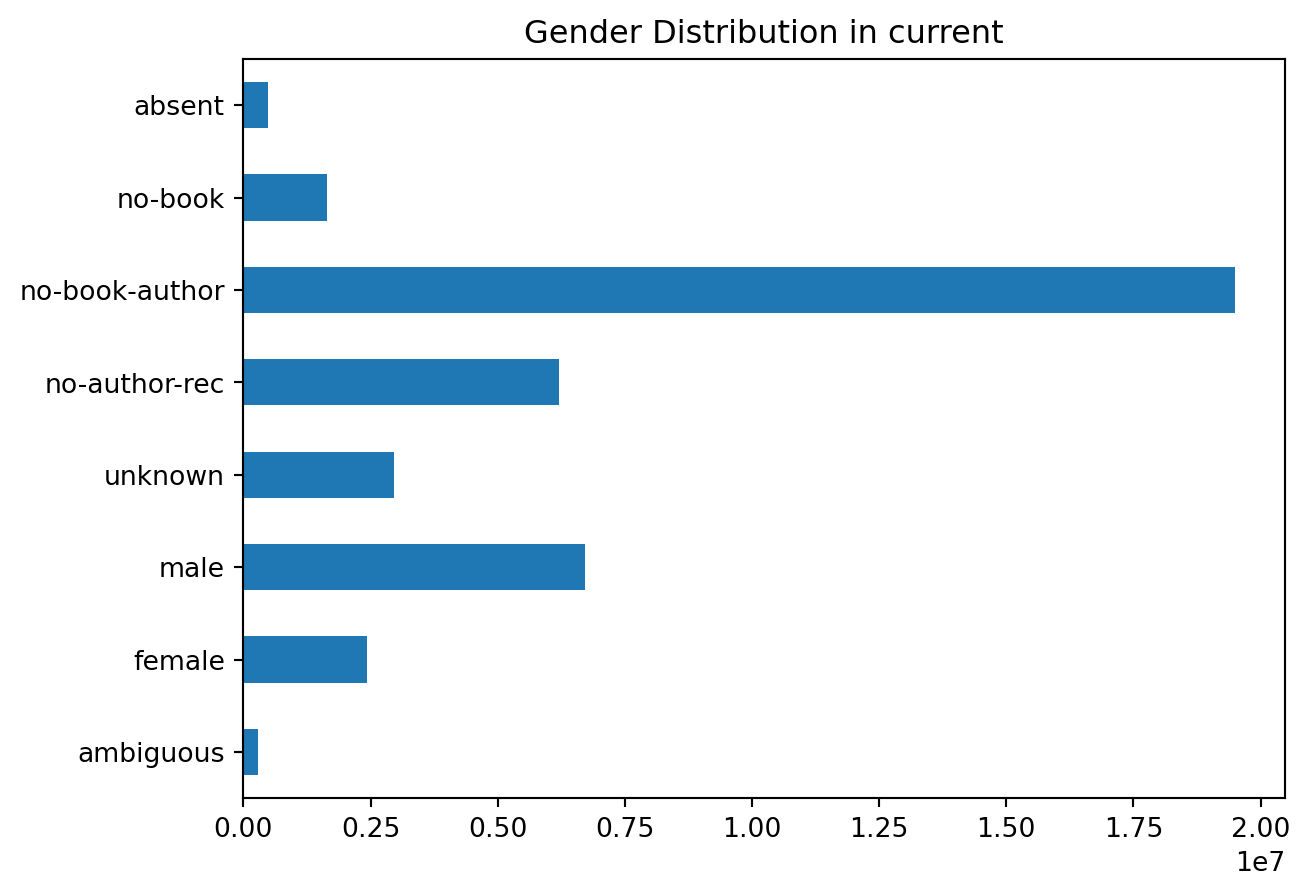
Release 3.0 (June 2025) to Current
Mostly technical code updates.
ct = gender_crosstab('2025-06-3.0', 'current')
ct.style.apply(style_row, 'columns')| current | ambiguous | female | male | unknown | no-author-rec | no-book-author | no-book | absent |
|---|---|---|---|---|---|---|---|---|
| 2025-06-3.0 | ||||||||
| ambiguous | 1.000000 | nan | nan | nan | nan | nan | nan | nan |
| female | nan | 1.000000 | nan | nan | nan | nan | nan | nan |
| male | nan | nan | 1.000000 | nan | nan | nan | nan | nan |
| unknown | nan | nan | nan | 1.000000 | nan | nan | nan | nan |
| no-author-rec | nan | nan | nan | nan | 1.000000 | nan | nan | nan |
| no-book-author | nan | nan | nan | nan | nan | 1.000000 | nan | nan |
| no-book | nan | nan | nan | nan | nan | nan | 1.000000 | nan |
| absent | nan | nan | nan | nan | nan | 0.955689 | nan | 0.044311 |
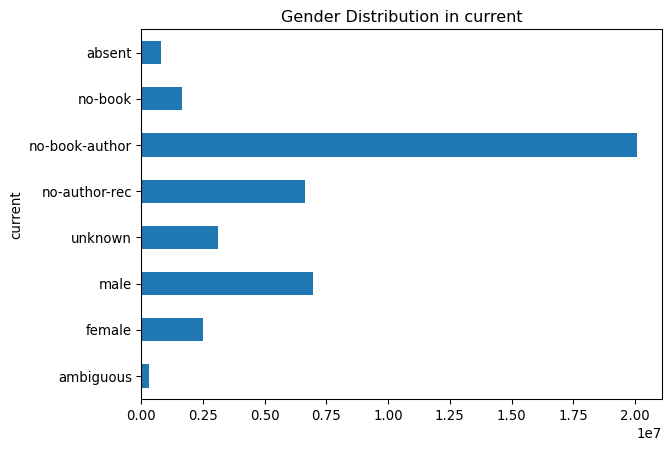
plot_gender('current')